Single-Cell Omics
Overview
Emerging Methods
Recent advances in instrumentation, biochemistry and computational tools for single-cell analysis have resulted in a rapid adoption of single-cell methods across different biological disciplines including cancer research and immunology. While single-cell RNA-sequencing is the most widely used technique to date, several promising methods for single-cell proteomics and single-cell multiomics are emerging. Performing high-throughput single-cell analysis on a routine basis requires robust instrumentation. At the same time, this highly dynamic field demands the use of flexible platforms that can accommodate a wide range of assays. Therefore, we provide an open, robust and automated platform for plate-based single-cell analysis workflows, including single-cell RNA sequencing, single-cell genome sequencing, single-cell proteomics as well as the latest assays for single-cell multiomics.


Image-based Cell Isolation
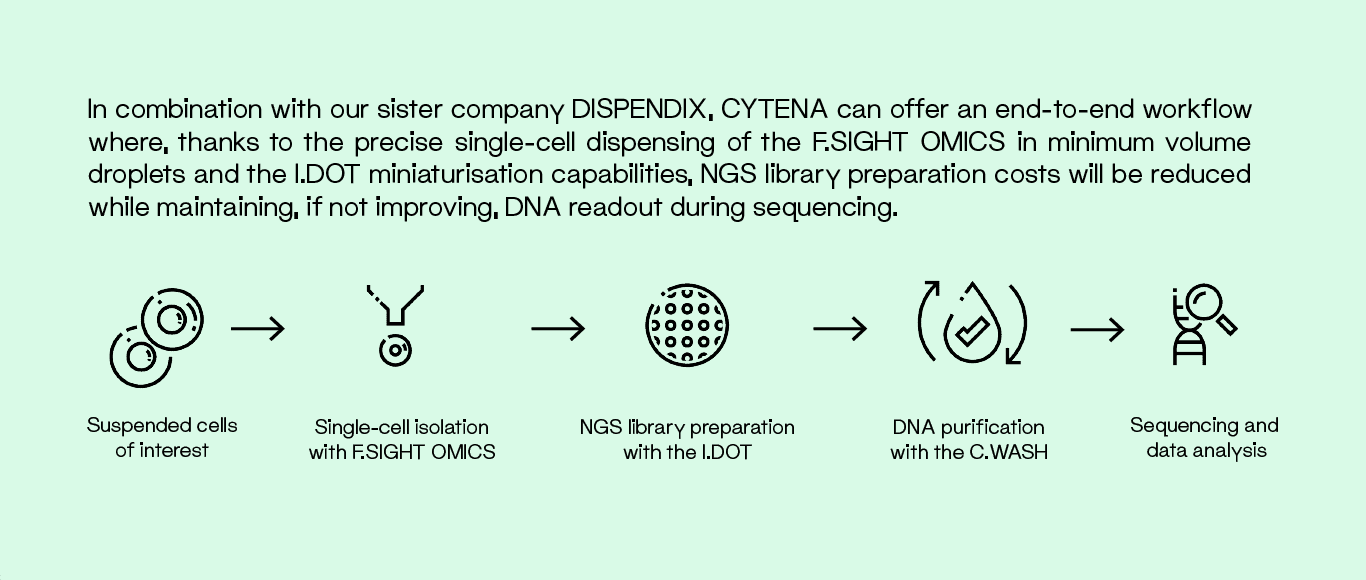
Our single-cell dispensing technology enables label-free cell isolation based on imaging. In addition, cells can be sorted based on fluorescent markers to exclude dead cells or target subpopulations. The cell isolation process is documented by a series of images that provide proof of successful single-cell isolation and insight into cell morphology such as size, roundness or fluorescent intensity. These parameters can be directly linked to the library size or the sequencing data.
Cell Lysis In Sub-microliter
Liquid Volumes
Accurate single-cell analyses and Next-Generation Sequencing (NGS) library preparation require efficient isolation of cells from the bulk sample and their precise deposition into PCR well plates. Our F.SIGHT OMICS is equipped with an automated offset correction (AOC) module that ensures cells are deposited into the center of each well, allowing cell lysis to occur in submicroliter volumes.
NGS Library Preparation
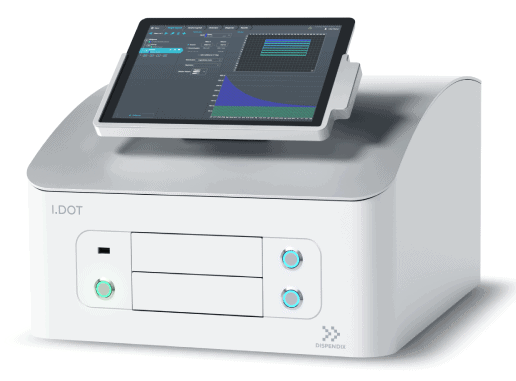
Automated liquid handling is key for routine single-cell library preparation as it reduces hands-on time and operator bias. Furthermore, more and more labs are turning to submicroliter reagent dispensing to reduce the costs of expensive reagents as the sample size and overall complexity grows. To meet the growing demand for miniaturized single-cell library preparation, we’ve created the I.DOT, our noncontact liquid handling platform which helps reduce costs, resources and the risk of cross contamination .
The I.DOT can dispense nanoliter droplets ranging from 8 nl to 50 nl. This flexibility enables miniaturization in many different chemistries for OMICS analysis. From the very start of the workflow, hundreds of libraries can be quantitated and normalized within minutes using the I.DOT, which allows for highly multiplexed dispensing of up to 96 reagents in parallel.
The very next steps would be library indexing and barcoding. The I.DOT’s noncontact dispensing technology was developed to perform even the most complex liquid handling tasks at unrivaled speed. Up to 2304 libraries can be uniquely barcoded with a single I.DOT source plate and 96 unique barcodes can be dispensed in under 60 seconds.

DNA Purification
Once NGS library preparation is finished, the libraries undergo DNA Purification. Traditionally, magnetic beads have been used to facilitate these washing steps. However, pipette-based methodologies are considered invasive and can result in significant bead loss. With our C.WASH platform, libraries can be purified and washed with noncontact centrifugal forces, reducing bead loss and avoiding unnecessary sample manipulation. Additionally, the C.WASH uses noncontact liquid dispensing technology to dispense all buffers onto the plates gently and without ever touching the DNA-covered beads.
Library Indexing and Early Barcoding
The ability to index NGS libraries with unique DNA barcodes provides the basis for high-throughput single-cell sequencing. For single-cell full-length transcript sequencing or whole exome sequencing, individual libraries are usually indexed after amplification and fragmentation. Other RNA-seq protocols, such as CEL-Seq2, make use of early barcoding and pooling of single cells, requiring the addition of hundreds of barcoded primers before reverse transcription. Any cross contamination between barcode primers must be avoided, since it would make proper demultiplexing impossible.
Sub-Applications
Precision medicine
Novel, innovative single-cell omics methods enable detailed characterization of pathways and cell type states associated with human diseases. Genetic characterization of single cancer cells and single-cell transcriptomics studies allow researchers to decipher intratumor heterogeneity, and thus to study processes involved in cancer progression and mechanisms of therapy response and resistance.
Characterization of cell type populations and identification of rare cell types
Single-cell omics approaches are widely used today to characterize the various cell types in the human body and to elucidate their functions. The study of immune cells is a prime example of how complex networks of highly specialized cell types with distinct functional states can be studied in detail using a single-cell approach.
Developmental studies
Single-cell methods provide researchers with valuable information on lineage and cell type identity and are thus well suited for studying developmental processes including cell-type differentiation in model organisms such as nematodes, zebrafish and mice.
Research Workflows