- PRODUCTS
-
UP.SIGHT™ NEW
Optimized For Proof Of Monoclonality, Colony Tracking, Confluency, & Titer Measurement -
F.SIGHT™ 2.0
Optimized For Rapid Dispensing of Fluorescent Cells -
C.SIGHT™ 2.0
Optimized for Powerful Dispensing of Unlabeled Cells -
B.SIGHT™
Optimized For Rapid Microbial Single-Cell Isolation and Cultivation -
F.SIGHT™ OMICS
Optimized For Single-Cell-Omics -
F.SIGHT™
Optimized For Affordability And Flexibility -
C.SIGHT™
Optimized for Affordable Cell Line Development - Help Me Choose
-
UP.SIGHT™ NEW
- APPLICATIONS
- RESOURCES HUB
- COMPANY
- SHOP
Accelerating microbiome research
Unlock unprecedented insights with single-cell
isolation of complex microbiome samples and
accelerate culturing of unknown species.
isolation of complex microbiome samples and
accelerate culturing of unknown species.
Overview
A critical step in isolating and studying microbes from complex communities is to efficiently separate single community members to obtain pure cultures. This is a complicated process due to the uneven distribution and high complexity of mixed microbial samples, such as those from human feces or from environmental sources like marine samples or wastewater. Traditionally, the most common methodologies to obtain pure cultures are either colony streaking on agar plates or dilution-to-extinction.
However, these methodologies are limited in the successful production of pure cultures because of the unknown composition and heterogeneous cell replication rates. Additionally, even though there has been an increase in microbiology studies – in particular microbiota studies – the workflow for modern anaerobic culture has not changed much over the last 60 years, thus limiting the throughput of colony isolation.
CYTENA’s B.SIGHT bridges the gap between the ambitions of microbiology and its limitations to give researchers the ability to analyze previously unculturable samples. Using high-resolution microscopy, the B.SIGHT captures the smallest bacteria in real time for more accurate and robust isolation and sorting.

Benefits
Microbiome research
Accelerate single-cell isolation and cultivate unknown species from complex microbiome samples. With these insights, new diagnostic techniques can be engineered for the medical field. Additionally, ecological and agricultural research will benefit from new findings and studies.

Accelerated
anaerobic isolation
and cultivation
Description: Single-cell isolation revolutionizes microbiology research by enabling rapid and efficient isolation of bacteria from complex microbiome samples.
Single-cell genomics uncultured bacteria
Unlock the secrets of microbial dark matter one cell at a time without the need for cultivation.
Research Workflows
Anaerobic isolation and cultivation of microbiota samples
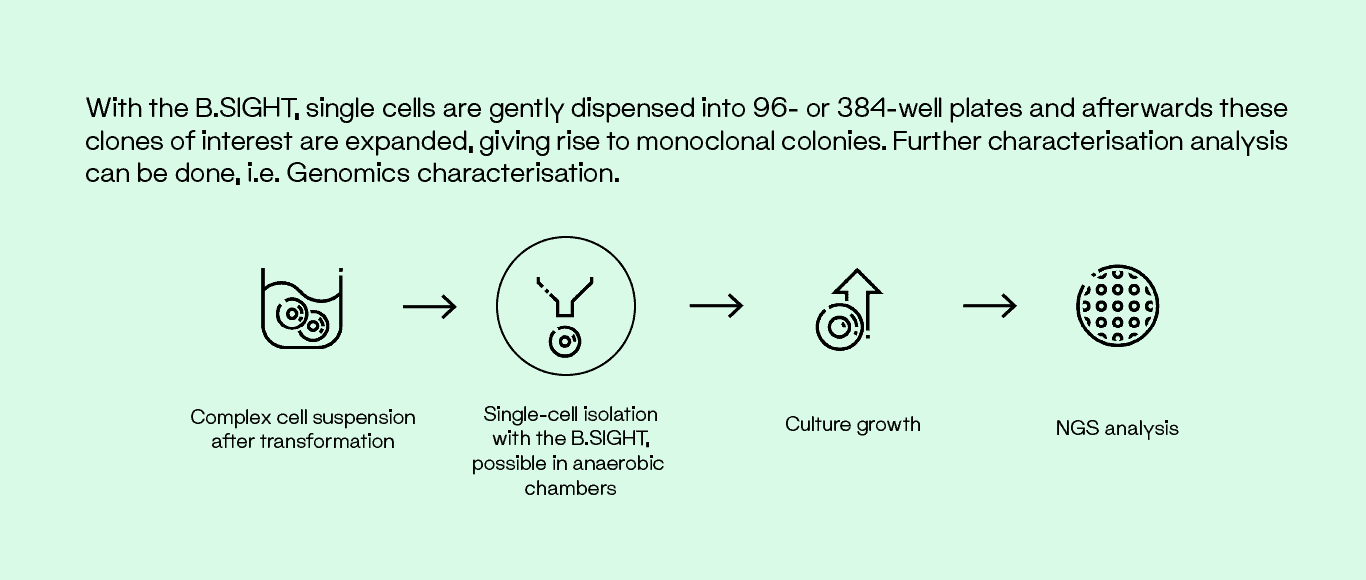
Cultivation using the classical agar plate method is used to study microbial communities. However, this method is time consuming and adds up to 21 days to go from microbiota to pure isolates. Single-cell isolation using the B.SIGHT allows high-throughput, label-free sorting of the smallest bacteria and accelerates the workflow by more than 5-fold.1 This device can efficiently separate cultures of both fluorescent and unlabeled bacteria, including complex samples.2
The B.SIGHT consists of a microfluidic system that constantly produces microdroplets of the sample buffer and uses high-resolution optical imaging and real-time analysis that guarantees dispensing of a single cell onto microwell plates. Droplets are formed through a non-pressure system and are dispensed onto the liquid culture or agar plates when they contain one single cell. If not, the droplets are discarded by an active vacuum system.
Sample preparation
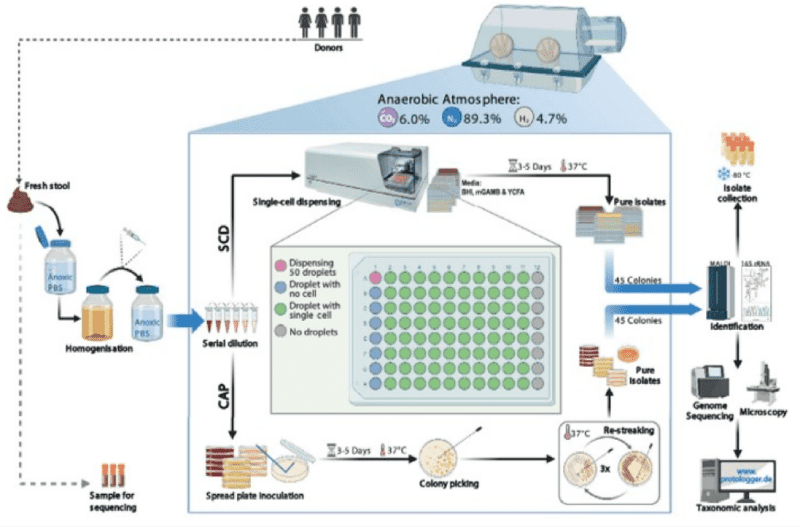
A microbiota sample, such as stool, can be suspended in anaerobic PBS supplemented with peptone (0.05% wt./vol.), l-cysteine (0.05% wt./vol.) and dithiothreitol (DTT) (0.02% wt./vol.) to obtain a 10-fold dilution (wt./vol.). The bottle containing the suspension must be flushed with an anaerobic gas mixture (89.3% N2, 6% CO2, 4.7 H2) prior to closing the lid. After vigorous manual shaking to re-suspend the sample and sterilizing the rubber stopper by flaming twice with alcohol, the faecal slurry can be diluted another 10-fold by rapidly transferred the mixture into another Schott bottle containing the same anaerobic medium using a syringe that has been flushed previously with the anaerobic gas mixture. The diluted anaerobic faecal dilutions can then be brought into the anaerobic workstation and further diluted with filter-sterilized anoxic PBS to a dilution of 10-8. The final dilution is filtered (0.2 μm) to remove any debris that may clog the nozzle of the B.SIGHT cartridge.2
The B.SIGHT can isolate and deposit a single cell per well in liquid culture media or directly onto solid agar. The initial sample preparation steps follow standard proceedure with a traditional culture and transformation. Afterwards, our workflow requires a simple cell suspension solution to be prepared to then be used in the B.SIGHT for single-cell isolation. Thanks to its small footprint, the device can be placed in an anaerobic chamber for sorting. Finally, the dispensed cells can be cultured and expanded in less time than with a traditional methodology.
Single-cell genomics of uncultivated bacteria
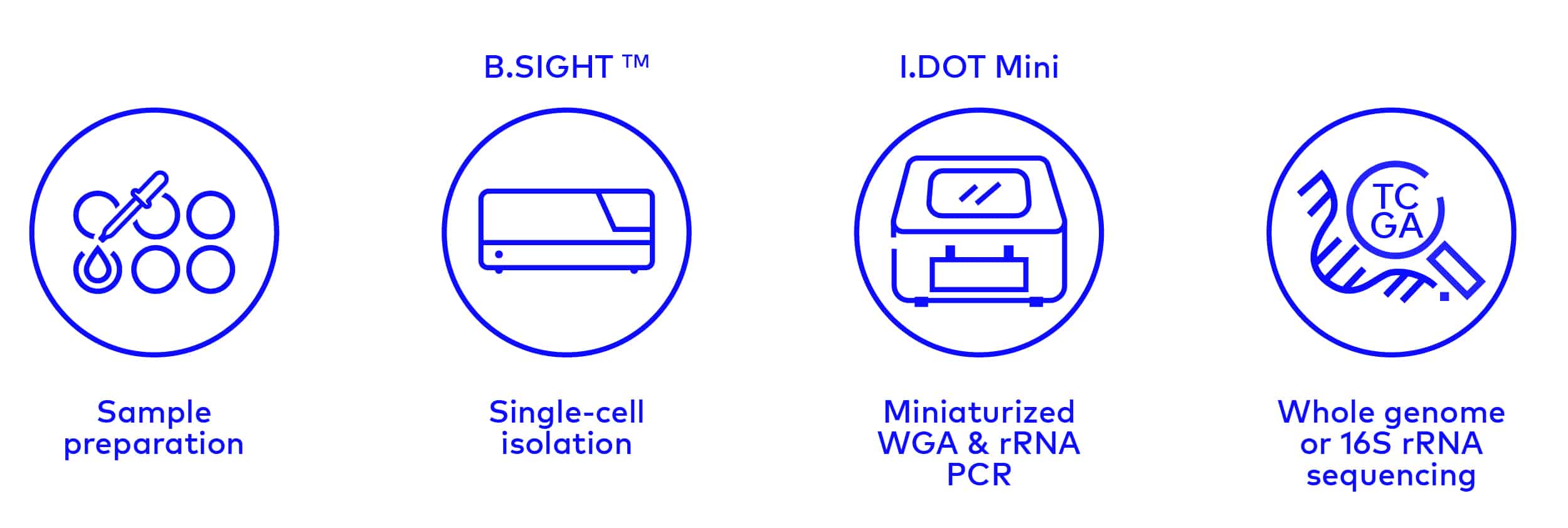
Most microbial organisms remain unexplored. Identifying new species holds enormous potential to discover new metabolic pathways that can be applied in biotechnological processes or be used to develop new therapeutics. Here, we present a workflow for exploring such microbial dark matter based on single-cell isolation and genetic characterization. The B.SIGHT can accurately dispense single microbial cells into PCR wells for subsequent whole genome amplification.
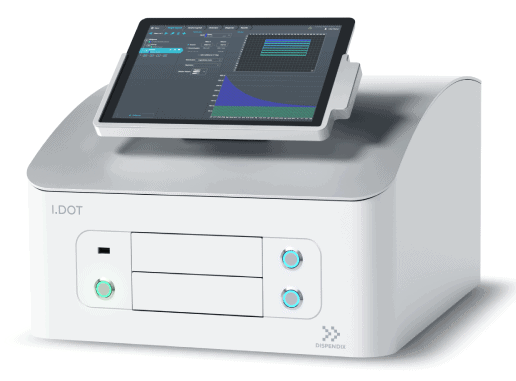
Thanks to the instrument’s ability to generate extremely small 50 pL droplets, the precise dispensing of single bacteria into PCR plates can be combined with the I.DOT, our contact-free low-volume liquid handling platform, to enable cell lysis in only 350 nL. This miniaturization scales down the amplification reaction by 20 folds, resulting in significant cost savings. Downstream sequencing of the 16S rRNA gene enables taxonomic classification of the isolated bacterial cells. Since all steps are performed in standard microwell plates, this workflow can easily be scaled up to analyze thousands of individual organisms to gain insight on the composition and heterogeneity of complex microbial samples without the need for cultivation.
Featured Resources
Previous
Next